-
Notifications
You must be signed in to change notification settings - Fork 110
2_2_step01_5
All steps can be sequentially processed using a batch script.
For more details of the commands, use -h option to see the usage.
Steps 0-3 to 0-5 are optional and can be run in no particular order.
LiCSBAS01_get_geotiff.py [-f FRAME] [-s start_date] [-e end_date] [--get_gacos] [--n_para int]
-f Frame ID (e.g., 021D_04972_131213). (Default: Read from directory name)
-s Start date (Default: 20141001)
-e End date (Default: Today)
--get_gacos Download GACOS data as well if available
--n_para Number of parallel processing (Default: 4)
This script downloads GeoTIFF files of unw (unwrapped interferogram) and cc (coherence) in the specified frame ID from COMET-LiCS web portal. The -f option is not necessary when the frame ID can be automatically identified from the name of the working directory. GACOS data can also be downloaded if available. Existing GeoTIFF files are not re-downloaded to save time, i.e., only the newly available data will be downloaded.
LiCSBAS02_ml_prep.py -i GEOCdir [-o GEOCmldir] [-n nlook] [-f frameID] [--n_para int]
-i Path to the input GEOC dir containing stack of geotiff data
-o Path to the output GEOCml dir (Default: GEOCml[nlook])
-n Number of donwsampling factor (Default: 1, no donwsampling)
-f Frame ID (e.g., 021D_04972_131213). Used only for downloading ENU
(Default: Read from directory name)
--n_para Number of parallel processing (Default: # of usable CPU)
This script converts GeoTIFF files of unw and cc to float32 and uint8 format, respectively, for further time series analysis, and also downsamples (multilooks) data if specified. Existing files are not re-created to save time, i.e., only the newly available data will be processed.
LiCSBAS03op_GACOS.py -i in_dir -o out_dir [-g gacosdir] [--fillhole] [--n_para int]
-i Path to the GEOCml* dir containing stack of unw data
-o Path to the output dir
-g Path to the dir containing GACOS data (Default: GACOS)
--fillhole Fill holes of GACOS data at hgt=0 in SRTM3 by averaging surrounding pixels
--n_para Number of parallel processing (Default: # of usable CPU)
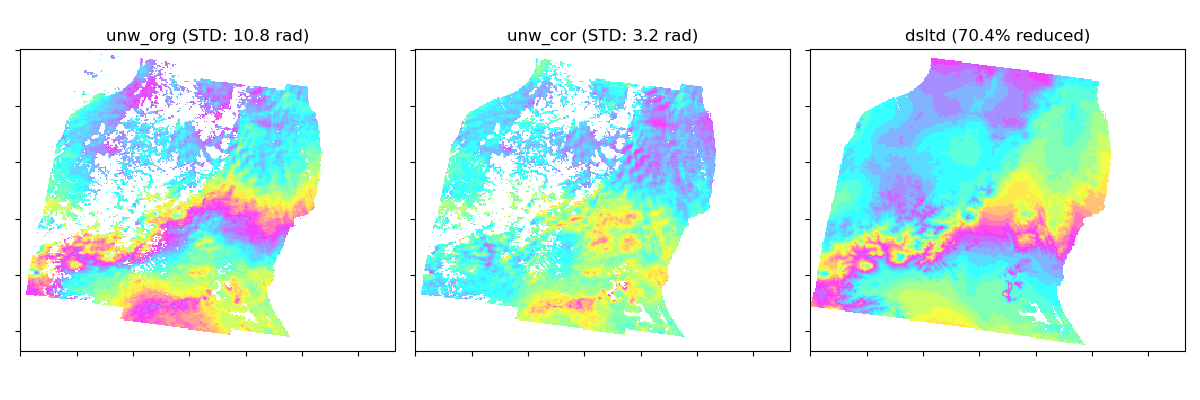
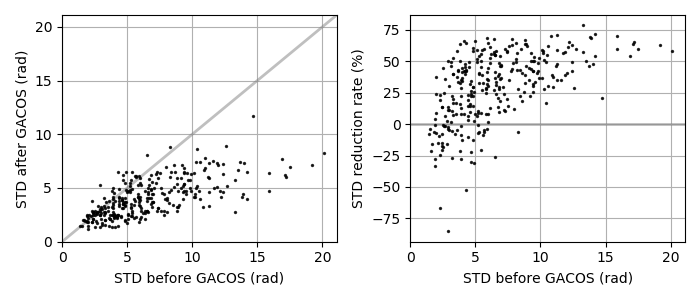
This script applies a tropospheric correction to unw data using GACOS data. GACOS data may be automatically downloaded from COMET-LiCS web at step01 (if available), or could be externally obtained by requesting on a GACOS web. If you request the GACOS data through the GACOS web, the dates and time of interest can be found in baselines and slc.mli.par, respectively. These are also available on the COMET-LiCS web portal. Once the GACOS data are ready, download the tar.gz, uncompress it, and put into GACOS dir. Existing files are not re-created to save time, i.e., only the newly available data will be processed. The impact of the correction can be visually checked by showing GACOS_info.png and */*.gacos.png. This step is optional.
LiCSBAS04op_mask_unw.py -i in_dir -o out_dir [-c coh_thre] [-r x1:x2/y1:y2] [-f txtfile] [--n_para int]
-i Path to the GEOCml* dir containing stack of unw data.
-o Path to the output dir.
-c Threshold for average coherence (e.g., 0.2)
-r Range to be masked. Index starts from 0.
0 for x2/y2 means all. (i.e., 0:0/0:0 means whole area).
-f Text file of a list of ranges to be masked (format is x1:x2/y1:y2)
--n_para Number of parallel processing (Default: # of usable CPU)
Note: either -c, -r or -f must be specified.
This script masks specified areas or low coherence areas in the unw data. The masking is effective when the unw data include areas which have many unwrapping errors and are not of interest, and can improve the result of Step 1-2 (loop closure). Existing files are not re-created to save time, i.e., only the newly available data will be processed. This step is optional.
LiCSBAS05op_clip_unw.py -i in_dir -o out_dir [-r x1:x2/y1:y2] [-g lon1/lon2/lat1/lat2] [--n_para int]
-i Path to the GEOCml* dir containing stack of unw data.
-o Path to the output dir.
-r Range to be clipped. Index starts from 0.
0 for x2/y2 means all. (i.e., 0:0/0:0 means whole area).
-g Range to be clipped in geographical coordinates (deg).
--n_para Number of parallel processing (Default: # of usable CPU)
This script clips a specified rectangular area of interest from unw and cc data. The clipping can make the data size smaller and processing faster, and improve the result of Step 1-2 (loop closure). Existing files are not re-created to save time, i.e., only the newly available data will be processed. This step is optional.