Table of Contents:
- Summary
- Requirements
- Installation
- Usage
- Combine output format
- Investigating event-level conservation
- Interconnection with VastDB web
- Interconnection with Matt
- Running as a container
- Issues
- Contributions
- Citation
- Species databases
- References
Vertebrate Alternative Splicing and Transcription Tools (VAST-TOOLS) is a toolset for profiling and comparing alternative splicing events in RNA-Seq data. It is particularly suited for evolutionary comparisons. It works synergistically with the VastDB web server, and Matt, a toolkit for downstream analyses of alternative splicing.
VAST-TOOLS requires the following software:
- bowtie 1.0.0 (Langmead et al., 2009)
- R 3.1 or higher, with the following packages installed (see Installation Section):
- optparse
- RColorBrewer
- reshape2
- ggplot2 >= v2.0
- MASS
- devtools
- psiplot
- Perl 5.10.1 or higher
- GNU coreutils
sort(all versions)
VAST-TOOLS also requires species-specific library files (collectively known as VASTDB), which must be downloaded separately. VASTDB consists of pre-made Bowtie indices, annotation, and template files. See below for installation instructions.
There are two components that need to be installed: VAST-TOOLS software package and VASTDB library files.
The VAST-TOOLS software package can be downloaded, unpacked and run as is. It does not require any additional installation steps. See Releases for the latest release or to get the latest development version, simply clone this repo:
> git clone https://github.com/vastgroup/vast-tools.git
VASTDB libraries must be downloaded separately and can be saved in the VAST-TOOLS
directory, or in an external location. If the latter, the path of VASTDB
must be supplied to vast-tools via --dbDir or alternatively, a symbolic
link can be created in the root of VAST-TOOLS directory. By default,
VAST-TOOLS looks for VASTDB inside its own directory
(e.g. ~/bin/vast-tools/VASTDB).
In addition to these libraries, VastDB also refers to a web server that provides information about AS events profiled by VAST-TOOLS through their stable VastID (e.g. HsaEX0040388). * NOTE: EventIDs are maintained across assemblies from the same species. While not all species are available in VastDB yet, they soon will.
These are the vast-tools assemblies available in v2.4.0:
1 - Homo sapiens (hg19, Hsa). Scaffold annotation: Ensembl v60.
2 - Homo sapiens (hg38, Hs2). Scaffold annotation: Ensembl v88.
3 - Mus musculus (mm9, Mmu). Scaffold annotation: Ensembl v62.
4 - Mus musculus (mm10, Mm2). Scaffold annotation: Ensembl v88.
5 - Bos taurus (bosTau6, Bta). Scaffold annotation: Ensembl v76.
6 - Gallus gallus (galGal3, Gg3). Scaffold annotation: Ensembl v65.
7 - Gallus gallus (galGal4, Gg4). Scaffold annotation: Ensembl v83.
8 - Xenopus tropicalis (xenTro3, Xt1). JGI_4.2. Scaffold annotation: Ensembl v84.
9 - Danio rerio (danRer10, Dre). Zv10. Scaffold annotation: Ensembl v80.
10 - Branchiostoma lanceolatum (braLan2, Bl1). Bl71nemr. Scaffold annotation: Ensembl Metazoa v46.
11 - Strongylocentrotus purpuratus (strPur4, Spu). Spur3.1. Scaffold annotation: SpBase.
12 - Drosophila melanogaster (dm6, Dme). Scaffold annotation: Ensembl Metazoa v26.
13 - Strigamia maritima (strMar1, Sma). Smar1. Scaffold annotation: Ensembl Metazoa v26.
14 - Caenorhabditis elegans (ce11, Cel). Scaffold annotation: Ensembl v87.
15 - Schmidtea mediterranea (schMed31, Sme). v31. Scaffold annotation: Dresden transcriptome & transdecoder.
16 - Nematostella vectensis (nemVec1, Nve). ASM20922v1/GCA_000209225.1. Scaffold annotation: Ensembl Metazoa v36.
17 - Arabidopsis thaliana (araTha10, Ath). TAIR10. Scaffold annotation: Ensembl Plants v31.
Automatic DB Installation:
You can automatically install the database files (and any pre-requisite R packages) using:
> ./install.R
Follow the command prompt to install automatically, and that should be it!
Of course you should add the vast-tools directory (e.g. ~/bin/vast-tools) to your PATH:
$ export PATH=~/bin/vast-tools:$PATH
$ echo 'export PATH=~/bin/vast-tools:$PATH' >> ~/.bashrc
Manual DB Installation:
For manual, install each species or all of them to any location by, e.g. for Hsa:
> wget http://vastdb.crg.eu/libs/vastdb.hsa.23.06.20.tar.gz
> tar xzvf vastdb.hsa.23.06.20.tar.gz
Available libraries and species (assembly, species_key):
Human (hg38, Hs2):
- Current version: vastdb.hs2.23.06.20.tar.gz (5.0G).
- Previous versions: vastdb.hs2.20.12.19.tar.gz (4.8G).
Human (hg19, Hsa):
- Current version: vastdb.hsa.23.06.20.tar.gz (6.3G).
- Previous versions: vastdb.hsa.20.12.19.tar.gz (6.3G), vastdb.hsa.16.02.18.tar.gz (6.3G).
Mouse (mm10, Mm2):
- Current version: vastdb.mm2.23.06.20.tar.gz (4.0G).
- Previous versions: vastdb.mm2.20.12.19.tar.gz (3.9G).
Mouse (mm9, Mmu):
- Current version: vastdb.mmu.23.06.20.tar.gz (5.7G).
- Previous versions: vastdb.mmu.20.12.19.tar.gz (5.7G), vastdb.mmu.16.02.18.tar.gz (5.7G).
Cow (bosTau6, Bta):
- Current version: vastdb.bta.23.06.20.tar.gz (3.4G).
- Previous versions: vastdb.bta.20.12.19.tar.gz (3.3G).
Chicken (galGal4, Gg4):
- Current version: vastdb.gg4.23.06.20.tar.gz (2.0G).
- Previous versions: vastdb.gg4.06.04.20.tar.gz (1.9G).
Chicken (galGal3, Gg3):
- Current version: vastdb.gg3.23.06.20.tar.gz (1.7G).
- Previous versions: vastdb.gg3.20.12.19.tar.gz (1.7G), vastdb.gg3.16.02.18.tar.gz (1.6G).
Xenopus (xenTro3, Xt1):
- Current version: vastdb.xt1.23.06.20.tar.gz (2.1G).
- Previous versions: vastdb.xt1.20.12.19.tar.gz (2.1G).
Zebrafish (Dre, danRer10):
- Current version: vastdb.dre.23.06.20.tar.gz (2.2G).
- Previous versions: vastdb.dre.20.12.19.tar.gz (2.2G), vastdb.dre.01.12.18.tar.gz (2.2G).
Amphioxus (braLan2, Bl1):
- Current version: vastdb.bl1.23.06.20.tar.gz (1.5G).
- Previous versions: vastdb.bl1.20.12.19.tar.gz (1.5G).
- Previous versions with species key "Bla" deprecated (a new assembly is being prepared for amphioxus).
Sea urchin (strPur4, Spu):
- Current version: vastdb.spu.23.06.20.tar.gz (1.2G).
- Previous versions: vastdb.spu.20.12.19.tar.gz (1.2G), vastdb.spu.01.12.18.tar.gz (1.2G).
Fruitfly (dm6, Dme):
- Current version: vastdb.dme.23.06.20.tar.gz (325M).
- Previous versions: vastdb.dme.20.12.19.tar.gz (319M), vastdb.dme.01.12.18.tar.gz (317M).
Centipede (strMar1, Sma):
- Current version: vastdb.sma.23.06.20.tar.gz (601M).
- Previous versions: vastdb.sma.20.12.19.tar.gz (585M), vastdb.sma.01.12.18.tar.gz (585M).
C. elegans (ce11, Cel):
- Current version: vastdb.cel.23.06.20.tar.gz (404M).
- Previous versions: vastdb.cel.20.12.19.tar.gz (395M), vastdb.cel.01.12.18.tar.gz (395M).
Planarian (schMed31, Sme):
- Current version: vastdb.sme.23.06.20.tar.gz (959M).
- Previous versions: vastdb.sme.20.12.19.tar.gz (952M), vastdb.sme.16.02.18.tar.gz (952M).
Sea anemone (nemVec1, Nve):
- Current version: vastdb.nve.23.06.20.tar.gz (692M).
- Previous versions: vastdb.nve.20.12.19.tar.gz (679M), vastdb.nve.01.12.18.tar.gz (679M).
Arabidopsis thaliana (araTha10, Ath):
- Current version: vastdb.ath.23.06.20.tar.gz (586M).
- Previous versions: vastdb.ath.20.12.19.tar.gz (568M).
** NOTE: from release v2.0.0, new VASTDB libraries are needed for all species.
** NOTE: release v2.3.0 included a new release of libraries (*.20.12.19.tar.gz). These libraries can be used (and are adviced to be used) with any previous vast-tools version.
** NOTE: EventIDs are maintained across assemblies from the same species.
If manually installed to central location, link the database files to vast-tools directory using:
> ln -s <path to VASTDB> VASTDB
If you did a manual install, you can test and see if you have everything that is necessary to run all of vast-tools, OR vast-tools will try and install R packages on the fly when necessary.
> ./install.R
If you did not use install.R as described above, you can manually install the
required R packages using the following commands:
> R -e 'install.packages(c("optparse", "RColorBrewer", "reshape2", "ggplot2", "devtools"))'
> R -e 'devtools::install_github("kcha/psiplot")'
VAST-TOOLS contains several main sub-commands: align, combine, merge, tidy, compare, diff, and plot. The following provides a quick introduction on how to run the tool:
Command usage can be retrieved through the -h (--help) flag to any sub-command:
> vast-tools [sub-command] -h
NOTE: Unless specified, all options are default, for example the output
directory is assumed to be 'vast_out', the database to be <path>/vast-tools/VASTDB.
To change these use the --output and -dbDir flags!
- NOTE: From v2.4.0, human is not the default species, which need to be provided.
VAST-TOOLS can be run as simply as:
> vast-tools align tissueA_rep1.fq.gz -sp hg38
> vast-tools align tissueA_rep2.fq.gz -sp hg38
> vast-tools align tissueB_rep1.fq.gz -sp hg38
> vast-tools align tissueB_rep2.fq.gz -sp hg38
> vast-tools combine -sp hg38
> vast-tools compare -a tissueA_rep1,tissueA_rep2 -b tissueB_rep1,tissueB_rep2
OR
> vast-tools diff -a tissueA_rep1,tissueA_rep2 -b tissueB_rep1,tissueB_rep2
> vast-tools plot INCLUSION-FILTERED.tab
You can speed up vast-tools significantly by allowing it to use multiple cores
by running it on a cluster. The -c flag can be passed to both align and
diff.
> vast-tools align tissueA_rep1.fq.gz -sp hg38 -c 8
AND
> vast-tools diff -a tissueA_rep1,tissueA_rep2 -b tissueB_rep1,tissueB_rep2
In this step, to increase the fraction of mapping junction reads within each RNA-Seq sample, each read is first split into 50-nucleotide (nt) read groups, using by default a sliding window of 25 nt (--stepSize option). For example, a 100-nt read would produce 3 overlapping reads (from positons 1-50, 26-75, and 51-100). In addition, both read mates from the paired-end sequencing are pooled, if available. For quantification, only one random count per read group (i.e. all sub-reads coming from the same original read) is considered to avoid multiple counting of the same original sequenced molecule. VAST-TOOLS align can also be used with pre-trimmed reads (--pretrimmed option), but only if reads have been trimmed by VAST-TOOLS. (Read headings need a special format so that they are properly recognized by VAST-TOOLS subscripts, and these are generated during the trimming process). Also, it is highly recommended that special characters ('-', '.', etc.) are not part of the fastq file names as this may cause unforeseen problems; use '_' instead. (The use of '-' is reserved for providing the read length (legacy) or specify the reads have been genome substracted; see below).
Next, these 50-nt split reads are aligned against a reference genome to obtain
unmapped reads, and these are then aligned to predefined splice junction libraries. Unmapped reads are saved
in the output directory as <sample>-50-e.fa.gz, where sample is the sample
name. The input reads can be compressed (via gzip) or uncompressed.
Currently, vast-tools supports multiple species and assemblies and is constantly growing in coordination with VastDB. From v2.4.0, the species is provided using the standard assembly (e.g. hg38, mm9, etc). The 3-letter species key, as in previous versions, can also be provided.
To enable gene expression analysis, use either the option --expr (PSI/PSU/PIRs pluscRPKM calculations [corrected-for-mappability Reads per Kbp and Million mapped reads; see Labbé et al, 2012 for details]) or --exprONLY (cRPKMs only). cRPKMs are obtained by mapping only the first 50 nucleotides of each read, or only the first 50 nucleotides of the forward read if paired-end reads are provided.
In addition to a file named *.cRPKM containing cRPKMs for each gene, a file name *.3bias will be created. This file contains information to estimate 3′ sequencing biases in the RNA-seq sample. Each of *.3bias file contains two rows:
- For all mRNAs with >200 mapped reads throughout at least 2500 nt, it provides the percentage of reads within the last five 500-nt windows, starting from the 3′-most window. The last column corresponds to the number of probed genes.
- For all mRNAs with >500 mapped reads throughout at least 5000 nt, it provides the percentage of reads within the last five 1000-nt windows, starting from the 3′-most window. The last column corresponds to the number of probed genes.
For example, to perform alignment with expression and 3′bias analysis on mouse data:
> vast-tools align mouse_tissue.fq.gz -sp mm10 --expr
If this alignment step needs to be repeated, the initial genome alignment step
can be skipped by supplying the <sample>-50-e.fa.gz file as input. VAST-TOOLS
will recognize the "-e.fa" suffix and start at the splice junction alignment
step. Gene expression and intron retention analyses cannot be run from this stage (you must start
from the raw reads).
> vast-tools align mouse_tissue-50-e.fa.gz -sp mm9
Although you can specify two fastq files to vast-tools in a 'paired-end' format,
the program treats both mates independently because of trimming, but will not double
count any trim or mate pair more than once (see above). Reads must be given to the program
such that vast-tools align fwd_mate_1.fq.gz rev_mate_2.fq.gz refers to two fastq
files of identical line number where Read1 from file_1 is mated to Read1 from file_2. NOTE: if reads are downloaded from SRA as sra files, use fastq-dump --split-file ./sample.sra to generate separate fastq files for each paired-end (plus a third file with unmatched mates).
Finally, from vast-tools v1.0.0 onwards it is possible to calculate intron retention using two slightly different approaches with the --IR_version 1/2 option. Version 1 is the one used in Braunschweig et al 2014. Version 2 incorporates alternative exon-exon junction for skipping reads to obtain a more representative Percent Intron Retention at the transcript level.
vast-tools merge can be used to pull the align outputs from various samples together into a new set of output files. This can be used to merge replicates when the read coverage for the independent replicates is not deep enough for a proper AS analysis. Unlike gene expression analysis, where 20-30 million reads is usually enough, differential AS analyses require deep sequencing coverage. Otherwise, only PSIs for AS events in highly expressed genes will be confidentely estimated, biasing the results. Since VAST-TOOLS uses only junction reads for quantification, we recommend at least 70 million reads per sample, and ideally >150 million reads. In our experience, the fraction of AS events whose PSI can be confidently estimated with VAST-TOOLS scales linearly with both read depth and length (specially if paired end) up to 200-300 million reads (depending on library complexity, read length, etc.). vast-tools merge is OPTIONAL; it is not needed in order to run vast-tools combine.
vast-tools merge needs a configuration file that is provided through the --groups option. This file must have two TAB-separated columns like this:
Sub_sample_A1 Sample_A
Sub_sample_A2 Sample_A
Sub_sample_A2 Sample_A
Sub_sample_B1 Sample_B
Sub_sample_B2 Sample_B
...
The subsample names must be identical to the first part of the file names of the RNAseq data before the first occurrence of a dot in the file name; usually everything until the file ending .fq.gz .
For paired-end RNAseq data, the subsample name is defined by the first RNAseq file used during the align step.
vast-tools merge will then merge the outputs of subsamples A1, A2 and A3 into a new Sample_A set of outputs that can then be used for combine.
> vast-tools merge --groups config_file
To merge cRPKM files from gene expression analysis, the option --expr needs to be provided. If so, vast-tools merge needs the species key (--sp) to upload the file with effective mappable positions per gene to recalculate cRPKMs. It is also possible to merge only cRPKM files by activating the option flag --exprONLY.
Finally, the subsample files can be moved to a subfolder (output_folder/PARTS) by using the option --move_to_PARTS.
From release v2.0.0, align recognizes automatically if RNAseq data are strand-specific and will align them strand-specifically.
Strand-unspecific RNAseq data will be aligned strand-unspecifically (equivalent to previous versions of VAST-TOOLS). This information will be gathered in a new file, SAMPLE.info, which will be generated together with the rest of outputs from align. This file will subsequently be used by other VAST-TOOLS modules. If the file is absent, VAST-TOOLS will assume an old version of VAST-TOOLS align was used and that the sample was thus mapped strand-unspecifically. Several samples with strand-specific/strand-unspecific RNAseq data can be merged into a new strand-specific/strand-unspecific sample with merge. Although it is also possible to merge samples with strand-specific and strand-unspecific RNAseq data into one new sample, it is not recommended to do so, as in this case the mappability correction will be applied in strand-unspecific mode which may introduce a bias to the final PSI/PIR values. It is possible to combine samples with strand-specific and strand-unspecific RNAseq data into a final output table with combine.
vast-tools combine will join all of the files sent to the same output directory found in <output_dir>/to_combine/, to form one final table in the main <output_dir> folder. This is the file you give to compare or diff in the case that you
intend to compare multiple samples. This output file contains a value for the percent of sequence inclusion (PSI/PSU/PIR) and a qual column for each sample. Details on the output format are provided below. In addition, the version of intron retention used in align can be specified using --IR_version (version 2 is used by default).
> vast-tools combine -o output_dir -sp [hg38|mm10|galGal4|etc] [Options]
From release v1.0.0-beta.3, it is possible to get the output in mm10 and hg38 coordinates. vast-tools will still work on mm9 and hg19, respectively, but all output coordinates are then lifted to the newer assemblies. This needs to be provided using the -a option.
From release v2.0.0, VAST-TOOLS includes a new module to identify and profile annotated exons (including constitutive exons). This is referred to as ANNOT, and it conceptually works as the splice-site based (aka COMBI) module (see Tapial et al, 2017 for details). Exons from the reference GTF annotation used to build VAST-TOOLS are quantified based on exon-exon junction reads and assigned a fixed ID (e.g. HsaEX6000001; exons from this module are labeled as "ANN" in the "COMPLEX" column of the output of combine, see below). Some annotated events are not present, as they are filtered for mappability and read imbalance. First and last exons are excluded. To obtain the legacy INCLUSION table, it is possible to use the option --noANNOT. NOTE: This module has not been as thouroughly tested and validated as the other exon skipping modules; therefore, lower validation rates for these events might be expected. This module requires new templates in VASTDB as well as an additional script (automatically provided in v2.0.0).
vast-tools provides two alternative modules (compare and diff) to perform differential splicing analyses on a reduced number of samples per group. Each module gives different functionalities.
compare: pre-filters the events based on read coverage, imbalance and other features, and simply compares average and individual dPSIs. That is, it looks for non-overlapping PSI distributions based on fixed dPSI cut-offs. For more than 3 replicates, it is likely to be too stringent.diff: performs a statistical test to assess whether the PSI distributions of the two compared groups are signficantly different. It is possible to pre-filter the events based on the minimum number of reads per sample, but subsequent filtering is highly recommended (e.g. overlapping the results with the output oftidy). For more than 5 samples per group it may also be over stringent.- When comparing multiple samples per group, an alternative approach is recommended. First, events should be pre-filtered using
tidy(see Simplifying Combine Table). This module allows to select events for which a minimum number of samples per group pass the quality controls. Then, a Mann-Whitney U-test or similar can be used to identify differentially spliced events. Finally, average dPSI per group should be calculated and a minimum difference (usually |dPSI| > 15) should be requested.
vast-tools compare identifies differentially spliced AS events between two groups (A and B) based mainly on the difference in their average inclusion levels (i.e. ΔPSI = average_PSI_B - average_PSI_A).
vast-tools compare takes any number of replicates for each group, provided either as sample names or column numbers (0-based) in the INCLUSION table. Then, it first filters out those AS events that do not have enough read coverage in ALL replicates (see Combine Output Format below for information on coverage thresholds). For intron retention, it is possible to filter out introns also based on a binomial test to assess appropriate balance of reads at the two exon-intron junctions by using --p_IR (see Braunschweig et al 2014 for details).
For valid AS events, vast-tools compare then requires that the absolute value of ΔPSI is higher than a threshold provided as --min_dPSI. In addition, it requires that the PSI distribution of the two groups do not overlap. This can be modified with the --min_range option, to provide higher or lower stringency. For example:
> vast-tools compare INCLUSION_TABLE.tab -a sampleA1,sampleA2 -b sampleB1,sampleB2 --min_dPSI 25 --min_range 5
will identify those AS events with an absolute ΔPSI between A and B higher than 25 and a minimum difference between the ranges of 5 (i.e. the maximum PSI value of group A and the minimum value of group B, if ΔPSI is positive, and the minimum PSI value of group A and the maximum value of group B, if ΔPSI is negative). --min_range can also take negative values to allow the PSI distributions of groups A and B to overlap to a defined extent (e.g. if --min_range -100, events will be called as differentially spliced as long as |ΔPSI| > N when --min_dPSI N).
By default, the comparison is not paired. A paired analysis can be done using the option --paired. If provided, each A replicate is compared with the corresponding one in group B. In this case, --min_dPSI is the minimum threshold for the average between each pair's ΔPSI, and --min_range is the minimum value any individual pair's ΔPSI can be (i.e. all individual ΔPSI's absolute values have to be larger than the value provided as --min_range and they all have to have the same sign [positive or negative]).
From v2.2.1, compare also requires that Alt3 (ALTA) and Alt5 (ALTD) events with differential splice site usage have a predicted significant overall impact in the transcript pool. Therefore, compare requires that the alternative splice sites being compared belong to an exon with a minimum inclusion level (~PSI) across ALL compared samples. This minimum PSI for the host exon is set to 25 by default, and can be modified using the --min_ALT_use option. The equivalent value to previous versions is --min_ALT_use 0. The PSI-like value is shown as the third value of the quality score (see Combine Output Format).
From v2.2.2, compare includes a new option, --noB3, to filter out EX events that have a very strong inclusion read imbalance. Specifically, if the exon has 0 reads supporting the inclusion from one side (upstream or downtream) and more than 15 in the other side in any of the compared samples, the exon is filtered out. These are exons that are most usually alternative first or last exons, but in some transcripts may behave as true cassette exons (i.e. they have a donor and an acceptor). The imbalance score for each exon and sample corresponds to the forth value of the quality score (see Combine Output Format).
Summary statistics are printed for a tally of AS events by type that show higher sequence inclusion in group A or B, the total number of events in vast-tools that have been profiled (i.e. that pass any applied filter), and the subset of the latter that are alternatively spliced in at least one of the compared samples (10<PSI<90).
Differentially spliced AS events are printed out to an output file. The name of this file is generated by default based on the parameters used, but it can also be provided using --outFile. The file is created in the directory of the input file. The selected events are then plotted automatically using vast-tools plot (see below), using a config file generated based on the type and order of the replicates provided as input. By default, all samples in the INCLUSION file are plotted, but it is possible to plot only the compared samples providing the --only_samples option. The plotting step can be skipped by activating the --no_plot flag.
It is also possible to output other sets of AS events of special interest for feature comparisons (e.g. using Matt). This can be done using the option --print_sets. It will produce three sets of AS events: (i) constitutive events (CS), which correspond to those with PSI < 5 (for IR) or PSI > 95 (for all other types) in all samples being compared; (ii) cryptic events (CR), which correspond to those with PSI > 95 (for IR) or PSI < 5 (for all other types) in all samples being compared; (iii) non-changing AS events (AS_NC), which correspond to those with 10 < av_PSI < 90 in at least one of the groups (or a range of PSIs > 10) and that do not change between the two conditions. The latter is specified with the option --max_dPSI. By default, it takes 1/5 of the provided --min_dPSI.
Finally, vast-tools compare can also produce list of gene IDs for the selected events to run Gene Ontology (GO) analyses using --GO. In particular, it generates four list: (i) differentially spliced cassette exons and microexons, (ii) introns with higher retention in B (IR_UP), (iii) introns with higher retention in A (IR_DOWN), and (iv) backgroup set, for all multiexonic genes that meet similar read coverage criteria. (The latter is crucial to avoid GO enrichment of highly expressed genes in the specific cell or tissue type of study). To generate the list of gene IDs, VAST-TOOLS needs to access VASTDB and thus needs the species key provided with -sp. Alternatively, a custom list of gene IDs for each AS event (event1\tgene_id1) can be provided using --GO_file, or gene symbols from the first colum of the INCLUSION table can be used instead activating the --use_names flag.
vast-tools diff provides functionality to test for differential AS based on
replicates and read depth for each event, but will also give reasonable estimates
if replicates are not available (Han, Braunschweig et al., 2017).
Bayesian inference followed by differential analysis of posterior distributions with respect to PSI/PSU/PIR. With replicate data, joint posterior distributions for a sample are estimated from empirical posterior distributions of the replicates using maximum-likelihood (MLE) fitting.
> vast-tools diff -a sampA_r1,sampA_r2,sampA_r3 -b sampB_r1,sampB_r2 -o outputdir -d outbase
Note: Sample names do not have to follow any specific convention as long as they remain valid ASCII words and any number of replicate sample names may be given 1:N.
Statistics Options
Probably the most important extra options to consider are -r PROB (--prob),
-m MINDIFF (--minDiff), -e MINREADS (--minReads), and -S MINSAMPLES (--minSamples)
These represent the stringency criterion for filtering of visual output and textual
data sent to file. -S is the minimum number of samples for each set -a and -b
that have to have at least -e reads each to be considered in the downstream
statistical comparison.
The -r flag represents the
minimal probability of acceptance that is required to consider a comparison to
be 'believable'. By default this is 0.95, but it can be altered depending on
stringency requirements.
The -m flag represents the minimum value of difference (MV, see example below)
between PSI in group A and PSI in group B that you will accept, such that we are are sure with at least
probability -r that there is a difference of at least -m. -m does not
currently alter the output sent to STDOUT, but does filter what is plotted to PDF
and printed to file.
The -e flag specifies the minimum number of reads for a sample/event to be
compared. In cases where the prior distribution has been methodically calculated
and/or is believable beyond an uninformative prior (like the uniform default),
this may not be necessary, however it is still highly recommended. The default
value for -e is 10, though this could easily be higher.
Additionally, diff allows you to alter the parameters of the conjugate beta
prior distribution, which is set as a uniform beta with shape parameters
--alpha and --beta as 1 and 1 respectively.
Beta shape parameters greater than one alter this probability distribution, and
may be more or less applicable to certain uses, see: beta
distribution
NOTE: When considering differential analysis of event types like intron retention
it may be more appropriate to use a custom prior model that is able to more accurately
reflect the lower expectation of inclusion levels.
In the case that you have paired samples, where NormalA is dependent on
PerturbationA, it is appropriate to use the --paired=TRUE flag. For
example when considering NormalA and NormalB, to compare to PerturbationA and
PerturbationB, the probability that P( joint_psi1 - joint_psi2 > -m ) is
calculated such that NormalA is only compared to PerturbationA, and then NormalB
is compared to PerturbationB. No MLE fitting is used in this case.
In all multireplicate cases where --paired=FALSE, the posterior distributions
of the individual replicates are used to estimate a 'best fit joint posterior' distribution
over PSI for each sample.
Performance Options
The -s flag can be used to specify the -s SIZE of the emperical
posterior distribution to sample, lower numbers decrease accuracy but increase
performance.
The diff command is also able to run in parallel. Specify the number of
cores to use with -c INT
Obviously more cores will increase the speed of diff, at the cost of increased
RAM usage.
Using the -n flag to specify the number of lines to read/process at a time,
will set a max threshold to the RAM used by parallel processing with the -c
flag. A lower number means that diff will use significantly less memory,
however by decreasing -n you have increased the number of times that the
mclapply function must calculate the parallel processing overhead. The
default is 100, which works well.
Output Format
The text output of diff looks like:
| GENE | EVENT | SampleA | SampleB | E[dPsi] | MV[abs(dPsi)]_at_0.95 |
|---|---|---|---|---|---|
| BOD1L | HsaEX0008312 | 0.124353 | 0.700205 | -0.575851 | 0.3 |
| KARS | HsaEX0032865 | 0.172134 | 0.460027 | -0.287892 | 0.22 |
| NISCH | HsaEX0043017 | 0.247743 | 0.500657 | -0.252915 | 0.09 |
| ALAS1 | HsaEX0003568 | 0.293333 | 0.537553 | -0.244220 | 0.09 |
| VPS13D | HsaEX0070518 | 0.984622 | 0.657589 | 0.327033 | 0.1 |
| TRO | HsaEX0067335 | 0.757929 | 0.474551 | 0.283378 | 0.04 |
| USP33 | HsaEX0069762 | 0.337669 | 0.845228 | -0.507560 | 0.14 |
| BCORL1 | HsaEX0007940 | 0.213452 | 0.500425 | -0.286973 | 0.05 |
Where for example the first event HsaEX0008312 in the BOD1L gene has multireplicate point estimate
for SampleA of 0.12 and 0.7 for SampleB. While this gives an expected value for the difference of
PSI (dPsi/ΔPSI) between SampleA and SampleB of -0.57, the minimum value (MV) for |ΔPSI| at 0.95 is
0.3, meaning that there is a 0.95 probability that |ΔPSI| is greater than 0.3. Use this value
to filter for events that are statistically likely to have at least a minimal difference of some
magnitude that you deem to be biologically relevant. The -m argument (default 0.1) provides a
lower bound for events that will be plotted to PDF and printed to file based on MV. As a cutoff,
the default is meant to provide a reasonably stringent baseline, however you could relax this if you
would rather view more events that may have significant but modest changes.
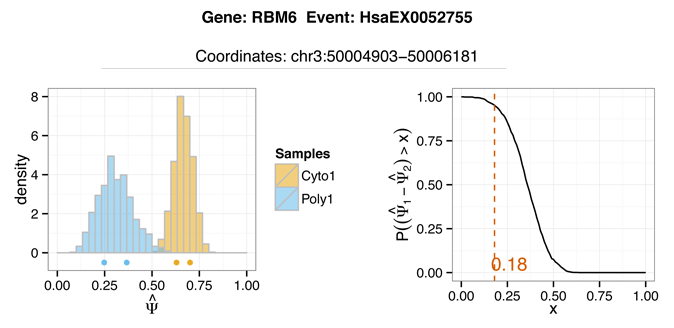
The output plot above shows in the left panel the two joint posterior distributions over PSI, and the
point estimates for each replicate plotted as points below the histograms. In the right panel,
the y-axis represents the probability of deltaPsi being greater than some magnitude value of x (shown on the x-axis).
The red line indicates the maximal value of x where P(ΔPSI > x) > -r, or the 0.95 default.
- NOTE: It is highly recommended that the output if further filtered (e.g. using
diff).
Using a similar logic to compare, vast-tools compare_expr identifies differentially expression genes between two groups (A and B) based on the difference in their average expression levels using the internal cRPKM metric. For this, it calculates the fold change in cRPKMs between the group averages (fold_B/A) as well as between each individual replicate. The default is set to a difference in fold change of the averages of at least 2 (--min_fold_av) and a difference between each of the individual replicates of 1.5 (--min_fold_r, equivalent to --min_range in compare). Paired comparisons are allowed using the option --paired. Basic usage:
> vast-tools compare_expr cRPKMS_AND_COUNTS-SpN.tab -a sample_a1,sample_a2 -b sample_b1,sample_b2 [options]
It requires an expression table with cRPKMs and read counts, which can be obtained in combine providing the option --C. vast-tools compare_expr performs several filters before doing the comparisons to discard lowly expressed genes across all samples or supported by too few reads overall. In particular, the default requires that all samples in at least one of the compared groups have a minimum cRPKM of 2. This can be modified using --min_cRPKM. Additionally, using --min_cRPKM_loose it is possible to allow only one sample across the comparison to have a minimum level of expression. With regards to the minimum number of reads to ensure an statistically sound cRPKM calculation, this is set to 50 by default, and can be modified using --min_reads.
By default, raw cRPKM values are compared. However, it is recommended the values are normalized. For this, the option --norm is provided, which using normalizeBetweenArrays from the limma R package. If this package is not installed in your computer, this can be done the first time you run vast-tools compare_expr by using the option --install_limma.
Finally, vast-tools compare_expr also provides an option to output files to perform Gene Ontology analyses (--GO). By defult, Ensembl GeneIDs are provided, but gene symbols can be retrieved instead using the option --use_names. The files provided are: (i) upregulated genes in B compared to A; (ii) downregulated genes in B compared to A; (iii) background set, after the expression and read count filters; (iv) the log2 (B/A fold change) values for all genes in the background set, which can be used for GSEA analyses.
VAST-TOOLS comes with a plotting script written in R. As of version 0.2.0, the core functionality of this script has been ported to the R package psiplot. Advanced users who would like to generate plots interactively in R are encouraged to use this package directly.
The input format follows the same format from the combine step described
above. The output is a pdf of scatterplots (one per AS event) of PSI values.
To execute from VAST-TOOLS, use the subcommand plot:
> vast-tools plot outputdir/significant_events.tab
It is recommended to filter the input file to a subset of events of interest
before plotting, such as those obtained from diff. Otherwise, the
resulting pdf file will be very large.
Plot customizations such as coloring and ordering of samples can be applied
using a configuration file. For more details on this advanced usage, see the
help description: vast-tools plot -h or see the
README of the psiplot
package. An example of sample input data and
configuration file template can be found under R/sample_data:
> vast-tools plot -c R/sample_data/sample_psi_data.config R/sample_data/sample_psi_data.tab
plot can also plot cRPKMs for gene expression by using the option --expr=TRUE. The options are similar as per AS plots, but you need to provide a cRPKM-only file (as generated in combine).
As per release v1.3.0, VAST-TOOLS comes with a script to simplify and filter the table obtained in combine, to make it more compatible with most R standard analyses. This module is called tidy, and it parses INCLUSION tables (from combine) event by event, printing out only the PSIs (i.e. no quality score) for those events that pass certain filters. Therefore, two main parameters need to be specified: (i) the minimum number of samples in which the event has sufficient read coverage (either as --min_N, absolute number of samples, or as --min_Fr, fraction of the total samples), and (ii) the minimum standard deviation of the PSIs of the events among the samples with good coverage (--min_SD). Several other parameters can be specified: --noVLOW, excludes samples with VLOW coverage; --p_IR, excludes samples that do not pass the binomial test for IR; --onlyEXSK, only EX events are considered. For any given event, samples that do not meet the mininum coverage cut-off will be assigned a PSI = NA. From v2.2.2, tidy can also filter out events using --noB3 and --min_ALT_use X, as described for compare.
tidy can also be run using a config file (--groups FILE), in which TWO groups of samples are provided using the following format:
Sample1\tGroupA
Sample2\tGroupA
Sample3\tGroupB
...
In this case, tidy will apply the defined filters to each group independently (and only to the samples listed in the config file). Both groups have to pass those filters. This option is useful if, for instance, the user needs to compare two groups with multiple samples using standard statistical tests. E.g. when comparing 50 patients vs 60 controls, the use may decide to run tidy with --min_N 10 and run a Mann-Whitney U-test on the filtered output in which at least 10 samples will have sufficient read coverage in each of the groups.
The output of combine is a tab-separated table with an entry (row) for each predefined alternative splicing event. For each event, there are six columns with basic information about it, and then a pair of columns for each sample from align that is combined.
- Column 1: Official gene symbol.
- Column 2: VAST-DB event ID. Formed by:
- Species identifier: Hsa (Human), Mmu (Mouse), or Gga (Chicken);
- Type of alternative splicing event: alternative exon skipping (EX), retained intron (INT), alternative splice site donor choice (ALTD), or alternative splice site acceptor choice (ALTA). In the case of ALTD/ALTA, each splice site within the event is indicated (from exonic internal to external) over the total number of alternative splice sites in the event (e.g. HsaALTA0000011-1/2).
- Numerical identifier.
- Column 3: Genomic coordinate of the alternative sequence.
- Column 4: Length of the alternative sequence. In ALTD/ALTA events, the first splice site within each event has a length of 0 nt, by definition.
- Column 5: Full set of genomic coordinates of the alternative splicing event.
- For EX: chromosome:C1donor,Aexon,C2acceptor. Where C1donor is the "reference" upstream exon's donor, C2acceptor the "reference" downstream exon's acceptor, and A the alternative exon. Strand is "+" if C1donor < C2acceptor. If multiple acceptor/donors exist in any of the exons, they are shown separated by "+". NOTE: The "reference" upstream and downstream C1/C2 coordinates are not necessarily the closest upstream and downstream C1/C2 exons, but the most external ones with sufficient support (to facilitate primer design, etc). If you wish to perform analyses of exon features and/or draw RNA binding maps, you are recommended to use Matt.
- For ALTD: chromosome:Aexon,C2acceptor. Multiple donors of the event are separated by "+".
- For ALTA: chromosome:C1donor,Aexon. Multiple acceptors of the event are separated by "+".
- For INT: chromosome:C1exon=C2exon:strand.
- Column 6: Type of event.
- S, C1, C2, C3: exon skipping (EX) events quantified by the splice site-based or transcript-based modules, with increasing degrees of complexity (based on Score 5 for a wide panel of RNA-seq samples; see below and Irimia et al. 2014 for further information).
- ANN: exon skipping (EX) events quantified by the ANNOTATION module. Their IDs also start by ≥ 6 (e.g. HsaEX6000001).
- MIC: exon skipping (EX) events quantified by the microexon pipeline.
- IR: intron retention event.
- Alt3: ALTA events.
- Alt5: ALTD events.
Then, for each combined sample, a pair of columns:
- Column 7: Estimated percent of sequence inclusion (PSI/PSU/PIR). PSI: percent spliced in (for EX). PSU: percent splice site usage (for ALTD and ALTA). PIR: percent intron retention (for INT).
- Column 8: Quality scores, and number of corrected inclusion and exclusion reads (qual@inc,exc).
-
Score 1: Read coverage, based on actual reads (as used in Irimia et al, Cell 2014). This is the only coverage score used by
compare,tidyandplot:- For EX (except microexon module): OK/LOW/VLOW: (i) ≥20/15/10 actual reads (i.e. before mappability correction) mapping to all exclusion splice junctions, OR (ii) ≥20/15/10 actual reads mapping to one of the two groups of inclusion splice junctions (upstream or downstream the alternative exon), and ≥15/10/5 to the other group of inclusion splice junctions.
- For EX (microexon module): OK/LOW/VLOW: (i) ≥20/15/10 actual reads mapping to the sum of exclusion splice junctions, OR (ii) ≥20/15/10 actual reads mapping to the sum of inclusion splice junctions.
- For INT: OK/LOW/VLOW: (i) ≥20/15/10 actual reads mapping to the sum of skipping splice junctions, OR (ii) ≥20/15/10 actual reads mapping to one of the two inclusion exon-intron junctions (the 5' or 3' of the intron), and ≥15/10/5 to the other inclusion splice junctions.
- For ALTD and ALTA: OK/LOW/VLOW: (i) ≥40/25/15 actual reads mapping to the sum of all splice junctions involved in the specific event.
- For any type of event: SOK: same thresholds as OK, but a total number of reads ≥100.
- For any type of event: N: does not meet the minimum threshold (VLOW).
-
Score 2: Read coverage, based on corrected reads (similar values as per Score 1).
-
Score 3: This score has been recicled to contain different information from release v2.2.2:
- EX (except microexon module): raw total read counts supporting upstream inclusion, downstream inclusion and skipping (format INC1=INC2=EXC).
- EX (microexon module): raw total read counts supporting inclusion and exclusion (format INC=EXC).
- ALTD and ALTA: PSI-like value of the exon hosting the ALTD/ALTA event. This score is used to filter out events in
comparebased on the option--min_ALT_use. - IR (from v2.1.3): corrected number of intron body reads (in a sample of 200bp in the middle of the intron, or the whole intron if shorter), and the number of mappable position in that sample (maximum 151 positions) (format READS=POSITIONS).
- Before v2.1.3: Read coverage, based on uncorrected reads mapping only to the reference C1A, AC2 or C1C2 splice junctions (similar values as per Score 1).
-
Score 4: This score has different meaning depending on the type of AS event:
- EX (except for microexon module): Imbalance of reads mapping to the inclusion splice junctions.
- OK: the ratio between the total number of corrected reads supporting inclusion for splice junctions upstream and downstream the alternative exon is < 2.
- B1: the ratio between the total number of corrected reads supporting inclusion for splice junctions upstream and downstream the alternative exon is > 2 but < 5.
- B2: the ratio between the total number of corrected reads supporting inclusion for splice junctions upstream and downstream the alternative exon is > 5 (but none is 0).
- B3: when the corrected reads for inclusion from one side is at least 15 and 0 for the other. Used to filter out events in
comparewhen the option--noB3is activated. - Bl/Bn: low (between 10 and 14)/no read coverage (between 1 and 9) for splice junctions supporting inclusion.
- EX (microexon module): "na" (no information provided).
- ALTD and ALTA: raw read counts for the specific splice site, for the all the splice sites of the event together (=total reads) and for those supporting skipping of the host exon. In versions early than v2.2.2: total reads for the event for all combinations or only for the reference acceptor (for ALTD) or donor (for ALTA).
- IR: raw read counts mapping to the upstream exon-intron junction, downstream intron-exon junction, and exon-exon junction in the format EIJ=IEJ=EEJ). In versions earlier than v2.2.2, corrected counts were shown instead of raw read counts.
- EX (except for microexon module): Imbalance of reads mapping to the inclusion splice junctions.
-
Score 5: This score has different meaning depending on the event type:
- EX (except for microexon module): Complexity of the event. The score refers to the number of reads that come from the "reference" C1A, AC2 and C1C2 junctions. Complexity increases as: S < C1 < C2 < C3.
- S: percent of complex reads (i.e. those inclusion- and exclusion-supporting reads that do not map to the reference C1A, AC2 or C1C2 splice junctions) is < 5%.
- C1: percent of complex reads is > 5% but < 20%.
- C2: percent of complex reads is > 20% but < 50%.
- C3: percent of complex reads is > 50%.
- NA: low coverage event.
- EX (microexon module): "na" (no information provided).
- ALTD and ALTA: similar complexity score as per exons. In this case, a given donor (for ALTA) or acceptor (for ALTD) is considered the "reference" site, and the complex reads are those coming from any other donor/acceptor.
- IR: p-value of a binomial test of balance between reads mapping to the upstream and downstream exon-intron junctions, modified by reads mapping to a 200-bp window in the centre of the intron (see Braunschweig et al., 2014).
- EX (except for microexon module): Complexity of the event. The score refers to the number of reads that come from the "reference" C1A, AC2 and C1C2 junctions. Complexity increases as: S < C1 < C2 < C3.
-
inc,exc: total number of reads, corrected for mappability, supporting inclusion and exclusion, such that PSI = inc/(inc+exc). Thus, when inclusion reads involve upstream and downstream junction sets, inc and exc are scaled.
-
A major feature of VAST-TOOLS event IDs is that they are fixed for each species (similar to Ensembl gene IDs, for example). This facilitates PSI comparisons across multiple samples and conditions (see next section) and evolutionary comparisons at the event level. Files with the correspondence for a given event ID between species are available for download in VastDB. For instance, for each event ID in Hsa, the corresponding orthologous event ID in Mmu (if available) can be found in this file. This unique feature allows to readily interconnect VAST-TOOLS results for different species.
VastDB is a web server that is tightly interconnected with VAST-TOOLS. VastDB contains information about AS events profiled by VAST-TOOLS for several species. It contains basic information about the events (including sequences, splice site strength, overlap with protein domains and disordered regions) and PSI quantifications for a large range of cell and tissue types and developmental stages, as profiled by VAST-TOOLS. Events have stable IDs that are identical in VAST-TOOLS and VastDB. Therefore, results obtained using VAST-TOOLS can be directly checked in VastDB to obtain physiological information about the AS events of interest. Moreover, VastDB provides event-level orthology information, allowing to compare information across the different species included in VastDB and VAST-TOOLS. Finally, general gene-level information is also provided, including quantifications of expression using cRPKMs.
Matt is a a toolkit for analyzing genomic sequences with focus on downstream analyses of AS events. It can be used to analyze the output of most tools to profile AS, but it has a specific module to facilitate the processing VAST-TOOLS tables.
It is possible to run VAST-TOOLS using software container technologies, simplifying so software installation and portability among different systems.
Example container setup with Docker:
docker run -d -v ~/myVASTDB:/VASTDB -v ~/myshared:/share --name myvast vastgroup/vast-tools tail -f /dev/null
vastgroup/vast-tools is the latest Docker image available. However, for production or publication purposes, we recommend to use always a specific version e. g.: vastgroup/vast-tools:v2.2.0
~/myVASTDB is the directory where VASTDB files are stored
~/myshared is a shared volume used for convenience for placing input and output files
The command highlighted above keeps the container running under the name myvast, so different VAST-TOOLS commands can be run from it.
Example command:
cd ~/myshared
fastq-dump SRR7802623
docker exec myvast vast-tools align /share/reads/SRR7802623.fastq -sp hg38 --expr -o /share/out/test
In HPC environments we strongly encourage to use Singularity. It is very easy to generate a Singularity image from Docker:
sudo /usr/bin/singularity build vast-tools.sif docker://vastgroup/vast-tools
/usr/bin/singularity exec ./vast-tools.sif vast-tools
Please report all bugs and issues using the GitHub issue tracker.
- Manuel Irimia (CRG)
- Andre Gohr (CRG)
- Nuno Barbosa-Morais (iMM)
- Ulrich Braunschweig (UofT)
- Kevin Ha (UofT)
- Tim Sterne-Weiler (UofT)
vast-toolsmain paper, including benchmarking and VastDB:
Tapial, J., Ha, K.C.H., Sterne-Weiler, T., Gohr, A., Braunschweig, U., Hermoso-Pulido, A., Quesnel-Vallières, M., Permanyer, J., Sodaei, R., Marquez, Y., Cozzuto, L., Wang, X., Gómez-Velázquez, M., Rayón, M., Manzanares, M., Ponomarenko, J., Blencowe, B.J., Irimia, M. (2017). An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res, 27(10):1759-1768
vast-toolsoriginal paper:
Irimia, M., Weatheritt, R.J., Ellis, J., Parikshak, N.N., Gonatopoulos-Pournatzis, T., Babor, M., Quesnel-Vallières, M., Tapial, J., Raj, B., O’Hanlon, D., Barrios-Rodiles, M., Sternberg, M.J.E., Cordes, S.P., Roth, F.P., Wrana, J.L., Geschwind, D.H., Blencowe, B.B. (2014). A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell, 59:1511-23.
- Intron retention analysis:
Braunschweig, U., Barbosa-Morais, N.L., Pan, Q., Nachman, E., Alipahani, B., Gonatopoulos-Pournatzis, T., Frey, B., Irimia, M., Blencowe, B.J. (2014). Widespread intron retention in mammals functionally tunes transcriptomes. Genome Research, 24:1774-86
diffmodule:
Han H, Braunschweig U., Gonatopoulos-Pournatzis T., Weatheritt R.J., Hirsch C.L., Ha K.C., Radovani E., Nabeel-Shah S., Sterne-Weiler T., Wang J., O'Hanlon D., Pan Q., Ray D., Vizeacoumar F., Datti A., Magomedova L., Cummins C.L., Hughes T.R., Greenblatt J.F., Wrana J.L., Moffat J., Blencowe B.J. (2017). Multilayered control of alternative splicing regulatory networks by transcription factors. Mol Cell, 65(3):539-553
- Chicken database:
Gueroussov, S., Gonatopoulos-Pournatzis, T., Irimia, M., Raj, B., Lin, Z.Y., Gingras, A.C., Blencowe, B.J. (2015). An alternative splicing event amplifies evolutionary differences between vertebrates. Science, 349:868-73
- Zebrafish and sea urchin databases:
Burguera, D., Marquez, Y., Racioppi, C., Permanyer, J., Torres-Mendez, T., Esposito, R., Albuixech, B., Fanlo, L., D'Agostino, Y., Gohr, A., Navas-Perez, E., Riesgo, A., Cuomo, C., Benvenuto, G., Christiaen, L.A., Martí, E., D'Aniello, S., Spagnuolo, A., Ristoratore, F., Arnone, M.I., Garcia-Fernàndez, J., Irimia, M. (2017). Evolutionary recruitment of flexible Esrp-dependent splicing programs into diverse embryonic morphogenetic processes. Nat Commun, 8:1799.
- Amphioxus, fruitfly, centipede, C. elegans and sea anemone databases:
Torres-Méndez, A., Bonnal, S., Marquez, Y., Roth, J., Iglesias, M., Permanyer, J., Almudí, I., O’Hanlon, D., Guitart, T., Soller, M., Gingras, A.-C., Gebauer, F., Rentzsch, F., Blencowe, B.J.B., Valcárcel, J., Irimia, M. (2019). A novel protein domain in an ancestral splicing factor drove the evolution of neural microexons. Nature Ecol Evol, 3:691-701.
- Planarian database:
Solana, J., Irimia, M., Ayoub, S., Orejuela, M.R., Zywitza, V., Jens, M., Tapial, J., Ray, D., Morris, Q.D., Hughes, T.R., Blencowe, B.J., Rajewsky, N. (2016). Conserved functional antagonism between CELF and MBNL proteins regulates stem cell-specific alternative splicing and regeneration in planarians. Elife, 5:e16797.
- Arabidopsis database:
Martín, G., Márquez, Y., Duque, P., Irimia, M. (2021). Alternative splicing landscapes in Arabidopsis thaliana across tissues and stress conditions highlight major functional differences with animals. Genome Biol, 22:35.
- Matt:
Gohr, A., Irimia, M. (2018). Matt: Unix tools for alternative splicing analysis. Bioinformatics, 35:130-132.
- cRPKMs:
Labbé, R.M., Irimia, M., Currie, K.W., Lin, A., Zhu, S.J., Brown, D.D., Ross, E.J., Voisin, V., Bader, G.D., Blencowe, B.J., Pearson, B.J. (2012). A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem Cells, 30:1734-45.
- bowtie:
Langmead, B., Trapnell, C., Pop, M., Salzberg, S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology, 10:R25.