getSpatialData is an R package in an early development stage that ultimately aims to provide homogeneous function bundles to query, download, prepare and transform various kinds of spatial datasets from open sources, e.g. Satellite sensor data, higher-level environmental data products etc. It supports both sf and sp classes as AOI inputs (see set_aoi in available functions). Due to the early development stage, the included functions and their concepts could be removed or changed in some cases.
For all public functions documentation is available. See also the list of data sources that are or will be implemented.
To install the current beta version, use devtools.
devtools::install_github("16EAGLE/getSpatialData")The following functions are publicly available and tested on Linux (Ubuntu 16.04 LTS, 17.10, 18.04 LTS) and Windows 10.
getSentinel_query()– querys the Copernicus Open Access Hubs for Sentinel-1, Sentinel-2 and Sentinel-3 data and returns a data frame containing the found records (rows) and their attributes (columns).getSentinel_restore()requests to restore Setninel datasets that have been archived by ESA to the Copernicus Long-Term Archive (LTA) (see argumentcheck_availofgetSentinel_query).getSentinel_preview()– uses the output ofgetSentinel_query()to preview (quick-look) a user-selected record even before downloading it. By default, the preview is displayed corner-georeferenced in a map viewer in relation to the session AOI.getSentinel_data()– uses the output ofgetSentinel_query()to download Sentinel data.
getLandsat_names()– obtains available Landsat product names from USGS Earth Explorer, which can be optionally used with getLandsat_query() to narrow the search.getLandsat_query()– querys USGS Earth Explorer for Landsat data and returns a data frame containing the found records (rows) and their attributes (columns).getLandsat_preview()– uses the output ofgetLandsat_query()to preview (quick-look) a user-selected record. By default, the preview is displayed corner-georeferenced in a map viewer in relation to the session AOI.getLandsat_data()– uses the output of getLandsat_query() to order and download Landsat data.- supports order (on-demand processing) and download of higher-level products (all Landsat products), e.g. top-of-atmosphere (TOA), surface reflectance (SR) or different indices, from USGS-EROS ESPA.
- supports direct download of Level-1 products (Landsat-8 only) via Amazon Web Services (AWS).
- will support direct download of Level-1 products (all Landsat products) via USGS EarthExplorer (requires a USGS user profile with machine-to-machine download permission)
getMODIS_names()– obtains available MODIS product names from USGS Earth Explorer, which can be optionally used with getMODIS_query() to narrow the search.getMODIS_query()– querys USGS Earth Explorer for MODIS data and returns a data frame containing the found records (rows) and their attributes (columns).getMODIS_preview()– uses the output ofgetMODIS_query()to preview (quick-look) a user-selected record. By default, the preview is displayed corner-georeferenced in a map viewer in relation to the session AOI.getMODIS_data()– uses the output of getMODIS_query() to order and download MODIS data from LAADS.
prepSentinel()beta – makes downloaded Sentinel datasets ready-to-use by automatically inspecting, extracting, sorting and converting the relevant contents of the datasets to a user-defined format.cropFAST()beta – crops a raster file to a spatial extent using GDAL. It is useful when working with large-scale, memory-intensive datasets.
login_CopHub()– define your Copernicus Open Access login credentials once for the present R session to be able to call eachgetSentinel*function without defining login arguments each time you use them.login_USGS()– define your USGS login credentials once for the present R session to be able to call eachget*function that connects to a USGS service without defining login arguments each time you use them.
set_archive()– define agetSpatialDataarchive directory to which all*_datafunctions will download data.set_aoi()- draw or define an AOI as sf, sp or matrix object for the running session that can be used by all query functions.view_aoi()- display the session AOI in an interactivemapview/leafletmap viewer.get_aoi()- get the session AOI you have defined or drawn before assf,spormatrixobject.
The following universal semantics on data are used by getSpatialData (from smallest to biggest entity):
image: An image of a specific time and spatial extent.record: A set of meta fields identifying and describing a specificimage, being part of multiple records in aquery.dataset: Smallest entity that is delivered by a service. Might consist of multiple files, including meta data and bandwise imagery. Covers a specific time and spatial extent.product: A data product offered by a specific service, consisting of multiple datasets over a period of time and a wide spatial extent. Might be differentiated by:platform: A general platform design (e.g. "Landsat" or "Sentinel").sensor: Type of sensor which acquired the data from which the product originates (e.g. "MODIS", "MSI" or "OLI").collection: A product version.level: Processing level of the product (e.g. "Level 2A" or "Surface Reflectance").source: The service acquiring, processing or distributing the product (e.g. "ESA Copernicus" or "USGS").
The following universal semantics on computational steps are used by getSpatialData:
get: Recieve data from different sources, named either bysensororplatform(whichever is used by the scientific community to referr to the derived products)names: Result of searching available products (differs bysourceandplatform), which might be differentiated further later on (e.g. bylevel).query: Result of searching asourcefor datarecordsof a specific or multipleproducts.preview: Preview arecord.data: Result of recieving one or multipledatasetfrom asource.
prep: Prepare/preprocess data obtained withget
The following code represents a working chain for querying, filtering, previewing and downloading Sentinel-2 data within R. The procedure can be done for Sentinel-1, Sentinel-2 or Sentinel-3.
## Load packages
library(getSpatialData)
library(raster)
library(sf)
library(sp)
## Define an AOI (either matrix, sf or sp object)
data("aoi_data") # example aoi
aoi <- aoi_data[[3]] # AOI as matrix object, or better:
aoi <- aoi_data[[2]] # AOI as sp object, or:
aoi <- aoi_data[[1]] # AOI as sf object
#instead, you could define an AOI yourself, e.g. as simple matrix
## set AOI for this session
set_aoi(aoi)
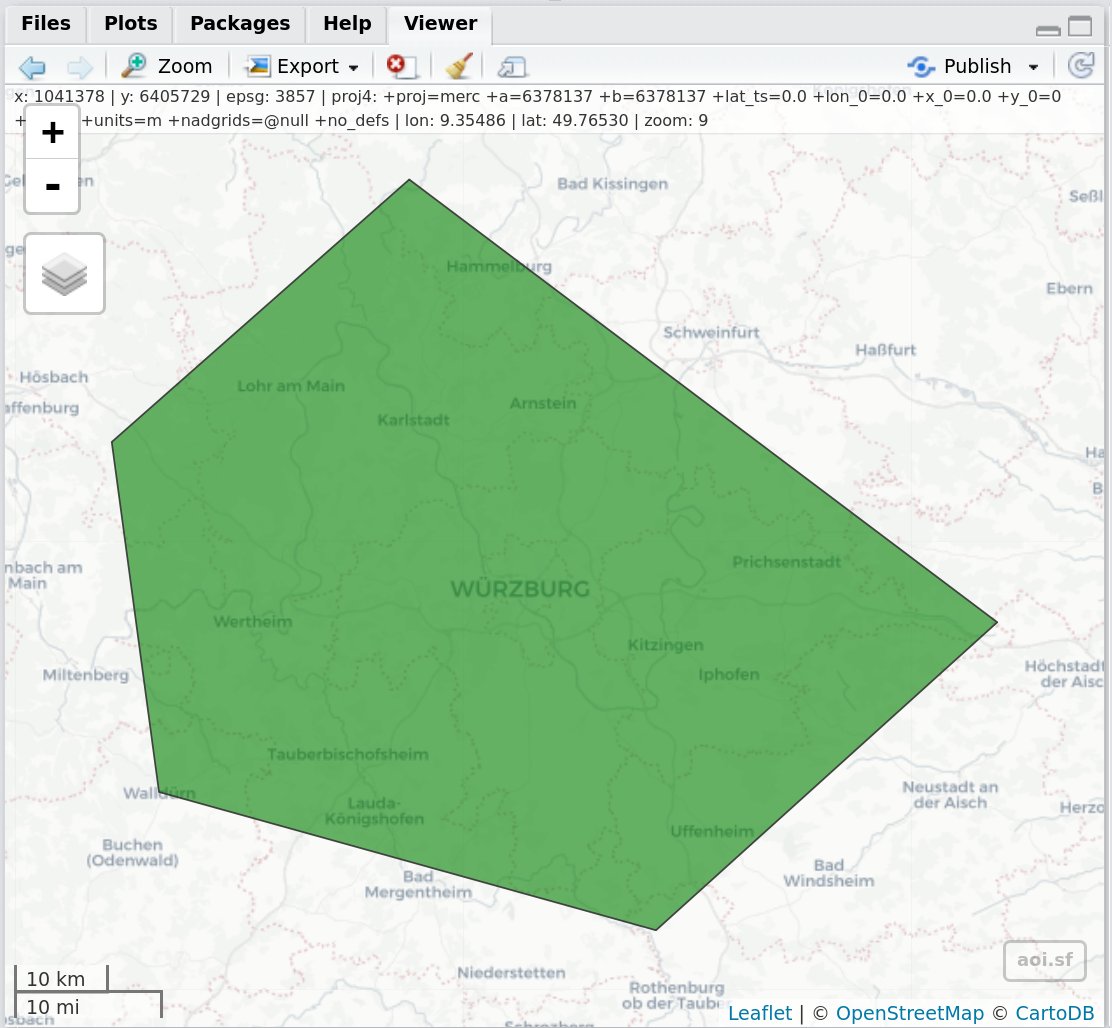
view_aoi() #view AOI in viewer, which will look like this:Figure 1: Screenshot of the RStudio Viewer, displaying the previously defined session AOI using view_aoi()
#instead of using an existing AOI, you can simply draw one:
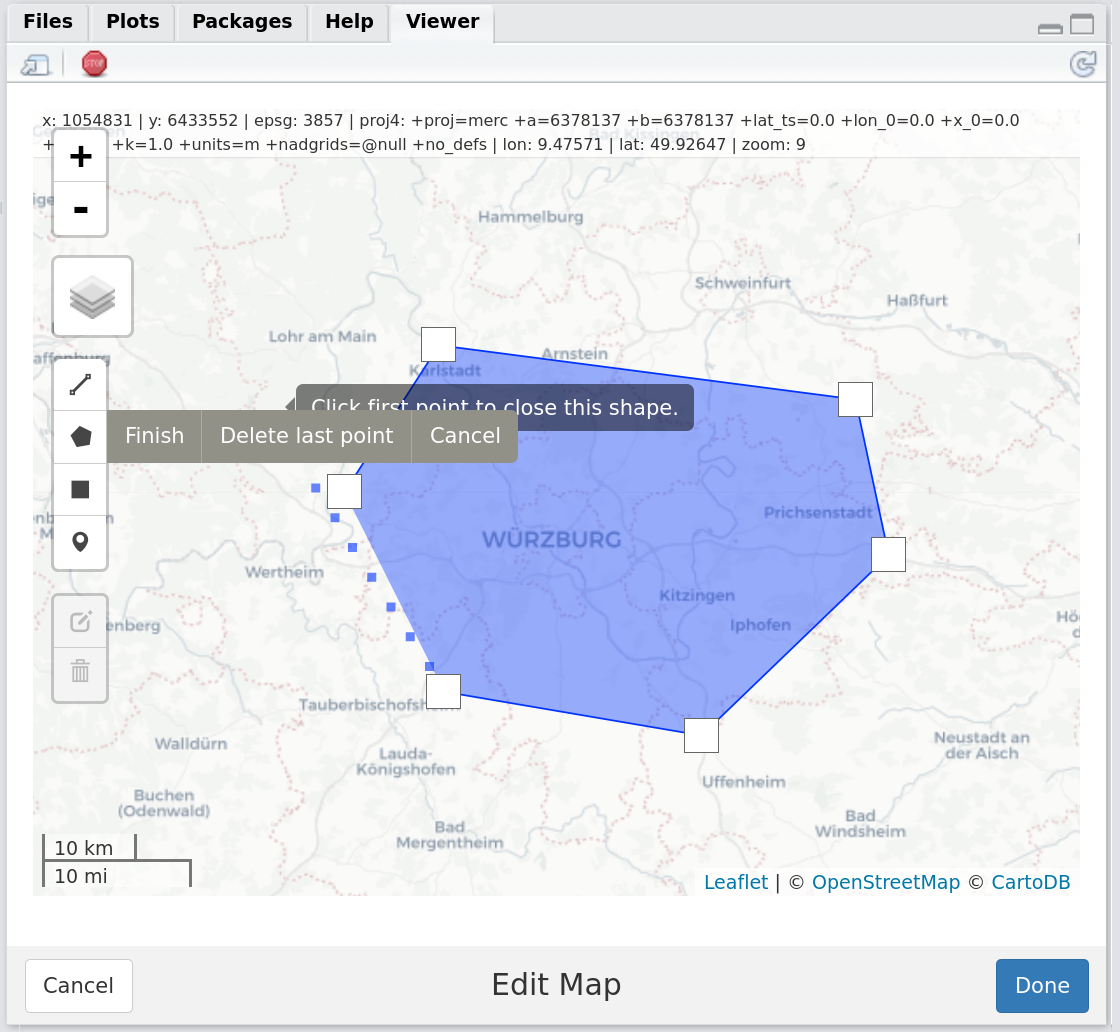
set_aoi() #call set_aoi() without argument, which opens a mapedit editor:Figure 2: Screenshot of the RStudio Viewer, displaying the mapedit editor allowing the user to draw a session AOI
## set login credentials and archive directory
login_CopHub(username = "your_username") #asks you for password
set_archive("/path/to/archive/")
## Use getSentinel_query to search for data (using the session AOI)
records <- getSentinel_query(time_range = c("2017-08-01", "2017-08-30"),
platform = "Sentinel-2") #or "Sentinel-1" or "Sentinel-3"
## Filter the records
colnames(records) #see all available filter attributes
unique(records$processinglevel) #use one of the, e.g. to see available processing levels
records_filtered <- records[which(records$processinglevel == "Level-1C"),] #filter by Level
records_filtered <- records_filtered[as.numeric(records_filtered$cloudcoverpercentage) <= 30, ] #filter by clouds
## View records table
View(records)
View(records_filtered)
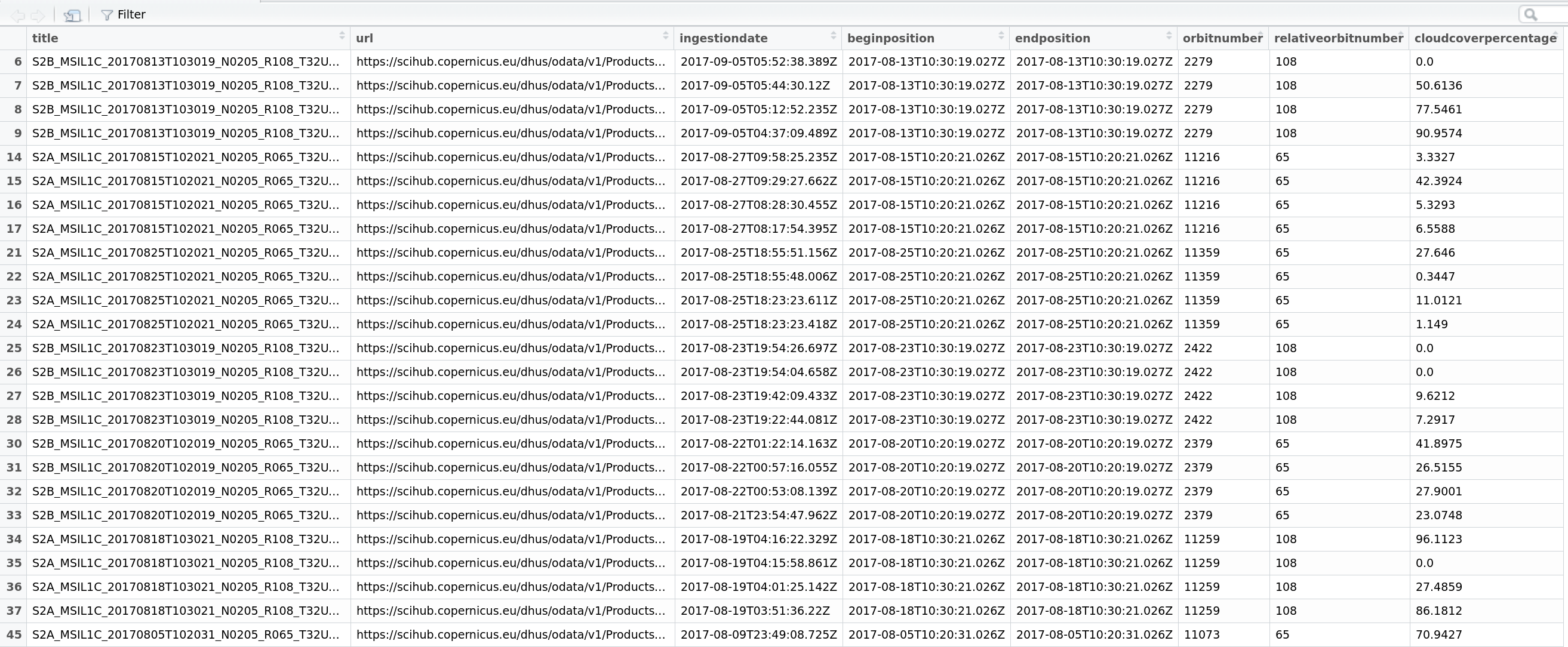
#browser records or your filtered recordsFigure 3: Screenshot of the View() display in RStudio, displaying a filtered records table produced by getSentinel_query()
## Preview a single record on a mapview map with session AOI
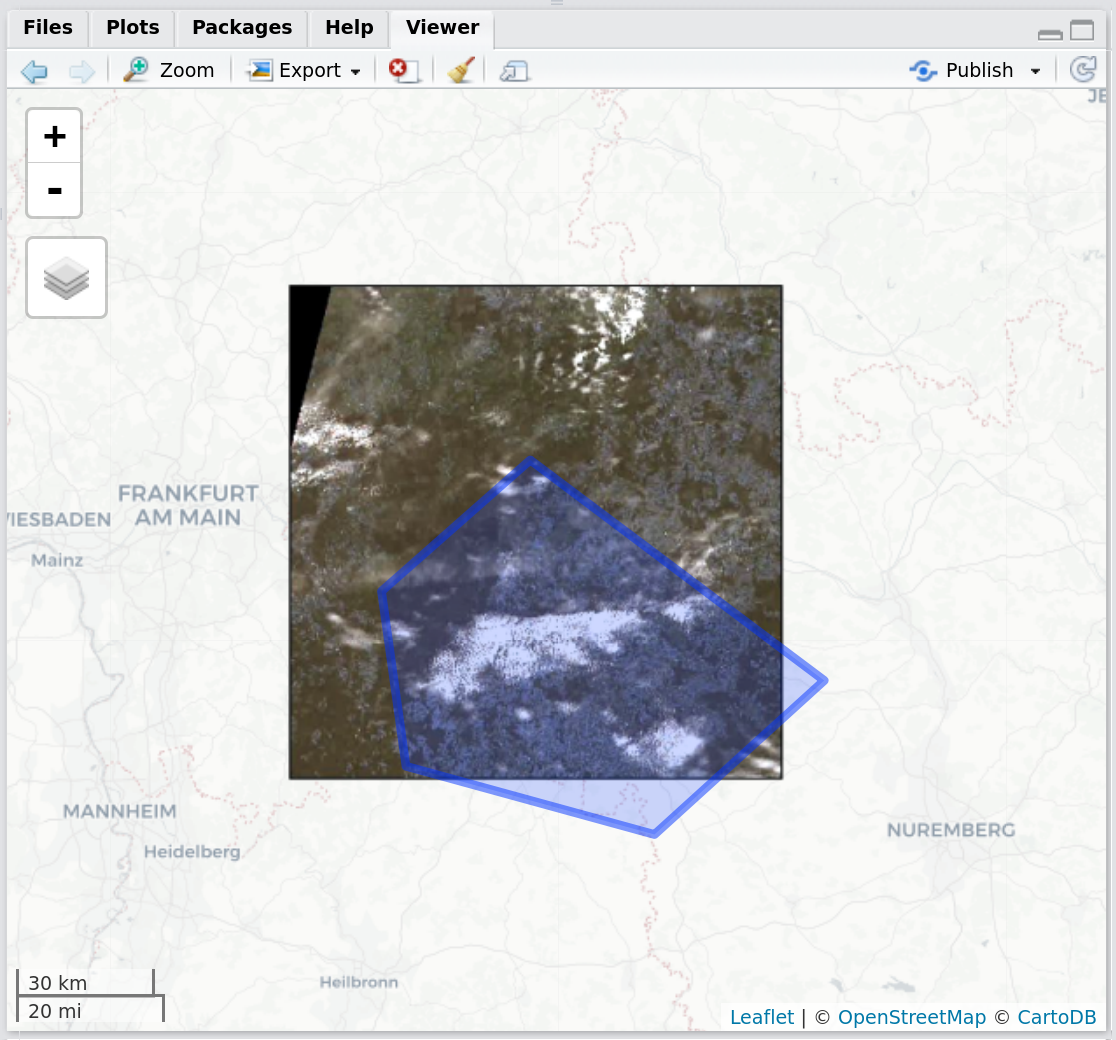
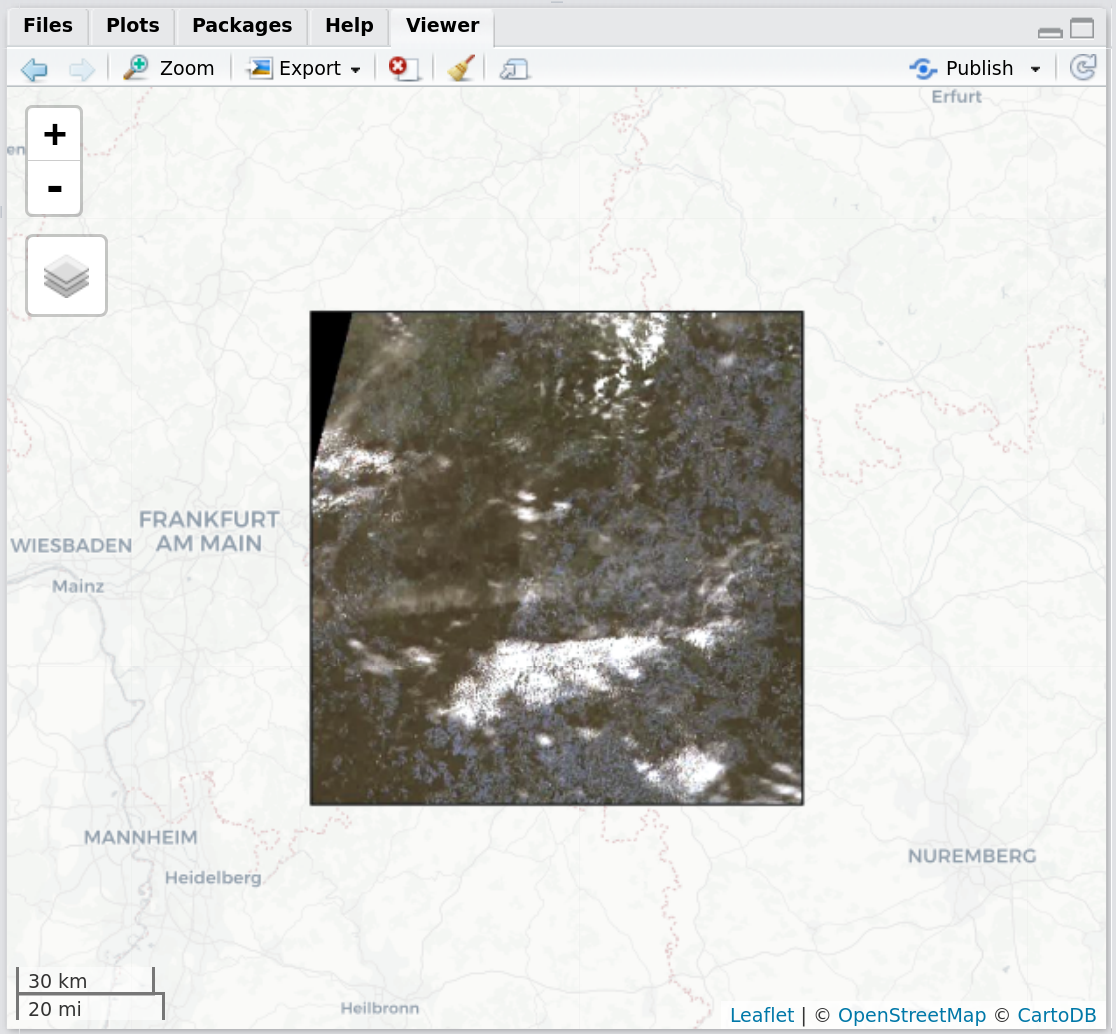
getSentinel_preview(record = records_filtered[9,])Figure 4: Screenshot of the RStudio viewer, displaying a corner-georeferenced Sentinel-2 preview and the session AOI using getSentinel_preview()
## Preview a single record on a mapview map without session AOI
getSentinel_preview(record = records_filtered[9,], show_aoi = FALSE)Figure 5: Screenshot of the RStudio viewer, displaying a corner-georeferenced Sentinel-2 preview using getSentinel_preview()
## Preview a single record as RGB plot
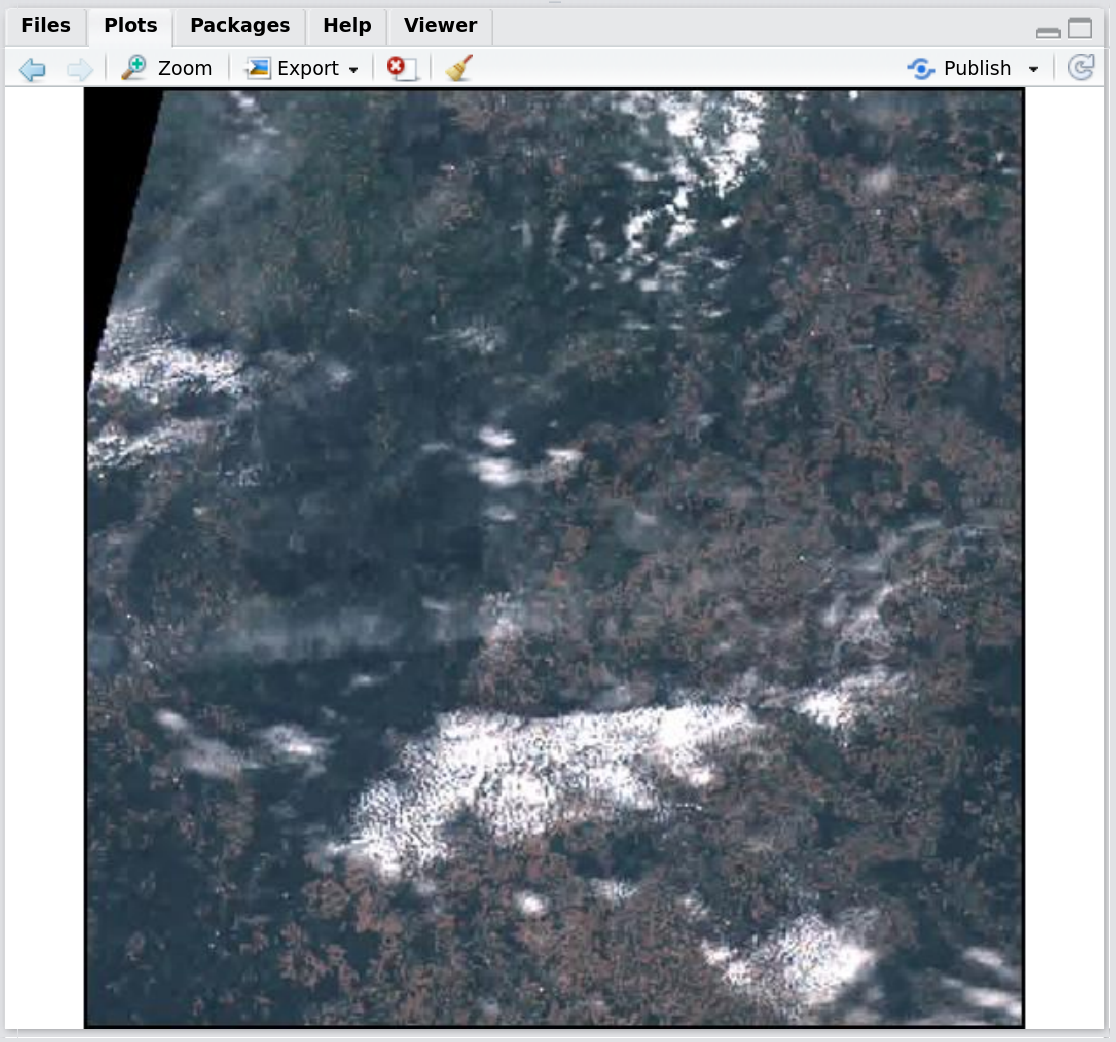
getSentinel_preview(record = records_filtered[9,], on_map = FALSE)Figure 6: Screenshot of the RStudio viewer, displaying a simple Sentinel-2 RGB plot preview using getSentinel_preview()
## Download some datasets to your archive directory
datasets <- getSentinel_data(records = records_filtered[c(4,7,9), ])
## Finally, define an output format and make them ready-to-use
datasets_prep <- prepSentinel(datasets, format = "tiff")
# or use VRT to not store duplicates of different formats
datasets_prep <- prepSentinel(datasets, format = "vrt")
## View the files
datasets_prep[[1]][[1]][1] #first dataset, first tile, 10 m resolution
datasets_prep[[1]][[1]][2] #first dataset, first tile, 20 m resolution
datasets_prep[[1]][[1]][3] #first dataset, first tile, 60 m resolution
## Load them directly into R
r <- stack(datasets_prep[[1]][[1]][1])
The following example shows how to query and then download MODIS imagery in parallel. This increases the overall download speed if enough bandwith is available to the client. The example has been contributed by Carina Kuebert.
## Load packages for working on multi-core
library(parallel)
library(doParallel)
library(foreach)
## getSpatialData
library(getSpatialData)
#### specify which files to download ####
# specify outdir (where files will be downloaded to)
outdir <- "/path/to/download/directory/"
# load example aoi
data("aoi_data")
set_aoi(aoi_data[[1]])
view_aoi()
# check, if service is available
services_avail()
## USGS login
login_USGS(username = "your_username")
# get available products
product_names <- getMODIS_names()
# query for records for your AOI, time range and product
time_range <- c("2019-01-01", "2019-01-10")
records <- getMODIS_query(time_range = time_range, name = grep("MOD09GA", product_names, value = T))
#### initiate cluster for paralell download ####
no_cores <- detectCores() - 1
cl <- makeCluster(no_cores, type = "PSOCK")
registerDoParallel(cl)
files <- foreach(i = 1:nrow(records[]),
.combine=c,
.packages='getSpatialData') %dopar% {
getMODIS_data(records[i, ], dir_out = outdir)
}
#### stop cluster ####
stopCluster(cl)The following products are being evaluated to be implemented within the package. This also includes sources which can be already accessed through existing packages that could be wrapped behind an standardized R function interface. Please feel free to contribute to the list, e. g. through a pull request:
Contribute! I'm happy about any kind of contribution, from feature ideas, ideas on possible data sources, technical ideas or other to bug fixes, code suggestions or larger code contributions! Open an issue to start a discussion: https://github.com/16eagle/getSpatialData/issues
getSpatialData has been mentioned here:
Kwok, R., 2018. Ecology’s remote-sensing revolution. Nature 556, 137. https://doi.org/10.1038/d41586-018-03924-9