This project, led by Matthew N. Tran and Kristen R. Maynard, describes a single nuclei RNA sequencing (snRNA-seq) project with data extracted from eight postmortem human brain donors collected by the Lieber Institute for Brain Development. Tran, Maynard and colleagues generated snRNA-seq data from five different brain regions:
- amygdala (AMY)
- dorsolateral prefrontal cortex (DLPFC)
- hippocampus (HPC)
- nucleus accumbens (NAc)
- subgenual anterior cingulate cortex (sACC)
The research findings derived from this dataset are described in the publications listed below. This data is also publicly available and is intended to serve as a resource for furthering our understanding of the transcriptional activity in the human brain. For example, the data from this study has been used by LIBD researchers for performing deconvolution of bulk RNA sequencing data. This resource is composed of 70,615 high-quality nuclei and you can download both the raw data as well as the processed data. Furthermore, you can explore interactively the data on your browser, make your custom visualizations, and export them to PDF files.
Finally, we have two versions of this resource. The initial version was shared on 2020 as a pre-print publication, while the peer-reviewed version was published in 2021. The pre-print version is limited as it contains data from three donors, while the peer-reviewed version was expanded to eight donors.
Matthew N. Tran, Kristen R. Maynard, Abby Spangler, Louise A. Huuki, Kelsey D. Montgomery, Vijay Sadashivaiah, Madhavi Tippani, Brianna K. Barry, Dana B. Hancock, Stephanie C. Hicks, Joel E. Kleinman, Thomas M. Hyde, Leonardo Collado-Torres, Andrew E. Jaffe, Keri Martinowich. Single-nucleus transcriptome analysis reveals cell-type-specific molecular signatures across reward circuitry in the human brain. Neuron 109, 3088-3103.E5. https://doi.org/10.1016/j.neuron.2021.09.001
Here's the citation information on BibTeX format.
@article {Tran2021,
author = {Matthew N. Tran and Kristen R. Maynard and Abby Spangler and Louise A. Huuki and Kelsey D. Montgomery and Vijay Sadashivaiah and Madhavi Tippani and Brianna K. Barry and Dana B. Hancock and Stephanie C. Hicks and Joel E. Kleinman and Thomas M. Hyde and Leonardo Collado-Torres and Andrew E. Jaffe and Keri Martinowich},
title = {Single-nucleus transcriptome analysis reveals cell-type-specific molecular signatures across reward circuitry in the human brain},
url = {https://doi.org/10.1016/j.neuron.2021.09.001},
year = {2021},
doi = {10.1016/j.neuron.2021.09.001},
publisher = {Elsevier {BV}},
volume = {109},
number = {19},
pages = {3088--3103.e5},
journal = {Neuron}
}
Matthew N. Tran, Kristen R. Maynard, Abby Spangler, Leonardo Collado-Torres, Vijay Sadashivaiah, Madhavi Tippani, Brianna K. Barry, Dana B. Hancock, Stephanie C. Hicks, Joel E. Kleinman, Thomas M. Hyde, Keri Martinowich, Andrew E. Jaffe. Single-nucleus transcriptome analysis reveals cell type-specific molecular signatures across reward circuitry in the human brain. bioRxiv 2020.10.07.329839; doi: https://doi.org/10.1101/2020.10.07.329839.
Here's the citation information on BibTeX format.
@article {Tran2020.10.07.329839,
author = {Tran, Matthew N. and Maynard, Kristen R. and Spangler, Abby and Collado-Torres, Leonardo and Sadashivaiah, Vijay and Tippani, Madhavi and Barry, Brianna K. and Hancock, Dana B. and Hicks, Stephanie C. and Kleinman, Joel E. and Hyde, Thomas M. and Martinowich, Keri and Jaffe, Andrew E.},
title = {Single-nucleus transcriptome analysis reveals cell type-specific molecular signatures across reward circuitry in the human brain},
elocation-id = {2020.10.07.329839},
year = {2020},
doi = {10.1101/2020.10.07.329839},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2020/10/08/2020.10.07.329839},
eprint = {https://www.biorxiv.org/content/early/2020/10/08/2020.10.07.329839.full.pdf},
journal = {bioRxiv}
}
We have provided 5 interactive websites that allow you to explore the data at single nucleus resolution for each of the brain regions. These interactive websites are powered by iSEE that allows you to add, hide, customize panels for visualizing the data. You can create any custom visualizations that you want and download both the code to make them as well as the figures you make. Please check the iSEE documentation for instructions on how to customize the panels. In particular, you might be interested in visualizing some of the marker genes from the lists provided below for the region-specific analyses.
- AMY: https://libd.shinyapps.io/tran2021_AMY/
- DLPFC: https://libd.shinyapps.io/tran2021_DLPFC/
- HPC: https://libd.shinyapps.io/tran2021_HPC/
- NAc: https://libd.shinyapps.io/tran2021_NAc/
- sACC: https://libd.shinyapps.io/tran2021_sACC/
If you want to make these websites on your own computer, check the shiny_apps directory.
If you are interested in exploring the data from the pre-print version which had three donors instead of eight, please check the following links.
- AMY: https://libd.shinyapps.io/tran2020_Amyg/
- DLPFC: https://libd.shinyapps.io/tran2020_DLPFC/
- HPC: https://libd.shinyapps.io/tran2020_HPC/
- NAc: https://libd.shinyapps.io/tran2020_NAc/
- sACC: https://libd.shinyapps.io/tran2020_sACC/
If you are interested in the raw data, that is the FASTQ files, they are publicly available from the Globus endpoint jhpce#tran2021 that is also listed at http://research.libd.org/globus/.
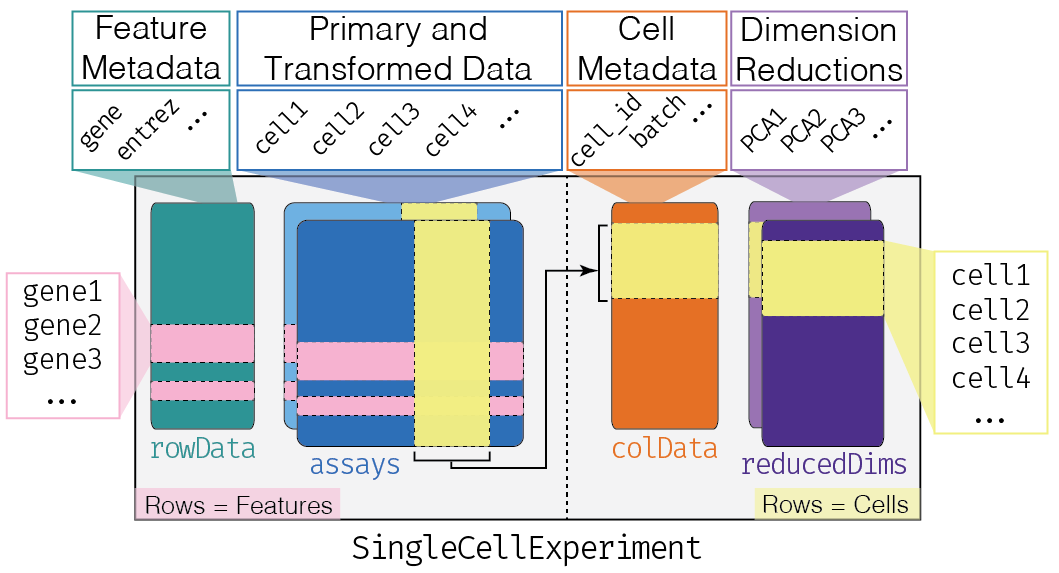
The corresponding SingleCellExperiment R/Bioconductor objects (with reducedDims, annotations, etc.) for each of the five regions across eight donors are publicly hosted at:
These files match the pre-print version that was composed of data derived from three donors.
If you are new to R/Bioconductor as well SingleCellExperiment objects, you might be interested in the:
- Orchestrating Single Cell Analysis (OSCA) with Bioconductor book
- 2020 WEHI scRNA-seq course taught by Peter Hickey with material adapted from OSCA
- following LIBD rstats club recorded sessions:
For more LIBD rstats club videos, check the following YouTube channel.
Here, nuclei are clustered and annotated within each brain region, separately, with markers defined at that level.
- AMY:
- DLPFC:
- HPC:
- NAc:
- sACC:
- AMY:
- DLPFC:
- HPC:
- NAc:
- sACC:
Here, nuclei are clustered across all brain regions, together, and then annotated, with markers defined at this level.
- Pre-print version:
- JHPCE location:
/dcs04/lieber/marmaypag/Tran_LIBD001/Matt/MNT_thesis/snRNAseq/10x_pilot_FINAL - FASTQ files:
/dcs04/lieber/lcolladotor/rawDataTDSC_LIBD001/raw-data/2021-10-15_Tran2021_published